
For academic visitors to our website, we provide on this page selected CompBioMed links and services of particular interest to you. Please check out the links on the column to the right, and stay tuned for more content. If you are interested in working more closely with us or visiting one of our partners for a collaboration, check out our Visitor Programme page for more information
In silico heart assays on High Performance Computing
Before new medicines reach patients several tests are done to detect possible risks and side effects. This is the reason why drugs are tested on millions of animals worldwide each year. But this landscape is changing: research shows computer simulations of the heart have the potential to improve the drug development process and to reduce the need for animal testing.
Drug development requires a long and costly pipeline, where the drug is tested in a gradually more realistic setting (i.e. the experimental model), where animals are the last step before human volunteers. The challenge is to identify the correct threads for human health, and not to miss the potential risk or value of a drug for example due to the differences between animals and humans.
In silico human models
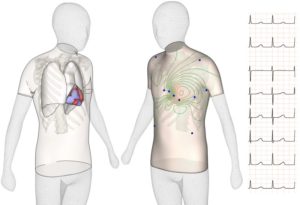 The revolutionary idea is to test a new drug in a “virtual human” organ, an experimental model entirely built on the circuits of a computer (i.e. in-silico). Recent research by the Computational Cardiovascular Science team at the University of Oxford demonstrates that computational models representing human heart cells show higher accuracy (89-96%) than animal models in predicting adverse drug effects such as dangerous arrhythmias – where the heart beat becomes irregular and can stop.
The revolutionary idea is to test a new drug in a “virtual human” organ, an experimental model entirely built on the circuits of a computer (i.e. in-silico). Recent research by the Computational Cardiovascular Science team at the University of Oxford demonstrates that computational models representing human heart cells show higher accuracy (89-96%) than animal models in predicting adverse drug effects such as dangerous arrhythmias – where the heart beat becomes irregular and can stop.
This research has recently been awarded with the International 3Rs Prize (from the National Centre for the Replacement Refinement and Reduction of Animals in Research) because of its potential to replace animal testing in labs. Instead of a one-model-fits-all method, the team uses an approach that simulates a wide range of responses under several conditions tested against experimental data. Everyone is different, and some drugs can have harmful side effects only for certain parts of the population, such as people with a specific genetic mutation or disease. This research also won the Technological Innovation Award at the Safety Pharmacology Society Meeting 2017.
This technology has been incorporated into a software, dubbed Virtual Assay, which is easy for non-experts to use in modelling and simulations. The software offers a simple user interface in which a control population of healthy cardiac cells with specific properties, based on human data, can be built. It can then be used to run in silico drug trials, before analysing the results. The whole process is very quick: it takes under five minutes using a modern laptop to test one drug in a population of 100 human cardiac cell models.
This research is part of a wider move towards the integration of computer models for drug safety testing which includes the Comprehensive in vitro Proarrhythmia Assay initiative, promoted by the US Food and Drug Administration and other organisations. The availability of high performance computing creates the possibility of running experiments with an unprecedented power, accounting for all the variability observed in the population and key related factors.
Pushing computer science boundaries
As part of the CompBioMed project we’re now working on 3D simulations of the heart to explore drug cardiac safety and efficacy on a larger scale. It also includes an exploration of diseased conditions, such as acute ischemia – where the blood flow in one of the arteries around the heart is obstructed. But while simulations of heart cells can run in a few minutes, 3D computer models of the whole heart still require a huge amount of computational power. The simulation of one heartbeat, for example, can take about three hours in a supercomputer with almost 1,000 processors.
The CCS team at the University of Oxford and the Barcelona Supercomputing Center have been awarded a PRACE Project Access on “In silico drug trials in the beating ischaemic human heart”, 2018-2020. This research is going to focus on developing a human electro-mechanical model of the heart using High Performance Computing (HPC) on the Marenostrum supercomputer. The project will conduct a proof of concept of a limited numbers of compounds and we will expand the investigations to consider a comprehensive list of medicines, building on the large datasets from our pharmaceutical industry collaborators. These models will be used to unravel key factors determining abnormalities caused by ischaemic disease and pharmacological therapy. Clinical imaging and electro-physiological dataset will be integrated to construct patient-specific anatomically –based electro-mechanical models of ischaemic human hearts, requiring tons of millions of nodes due to numerical constraints.
This research is part of a bigger movement towards the use of the virtual human and is being supported by the CompBioMed European project, an NC3Rs Infrastructure for Impact award, the TransQST European project, the PRACE project, an ARCHER project and the PIC project.
More examples of academic users:
- CompBioMed e-Seminar #31 - CompBioMed’s 31st e-seminar took place on 26 July 2023, titled “Assessing the Credibility of Computational Models: Application of the FDA-Endorsed ASME VV-40“. Computational models in the medical field are more and more stepping out the door of the laboratories they were developed in to find practical applications in the clinical or regulatory practice, for instance.…
- CompBioMed Conference 2023 – Abstract Submission Deadline Extended to 30 June 2023 - Due to popular demand, the abstract submission deadline for CompBioMed Conference 2023 has been extended to 30 June 2023. You can read more and submit abstracts on the conference website.
- CompBioMed Newsletter Issue 12 – Out Now - The latest issue of the CompBioMed Newsletter is now available, addressing the upcoming CompBioMed Conference 2023, the book Virtual You, the CompBioMed film Virtual Pandemics, our scalability service, and biomedical urgent computing in the exascale era. Click here to view it. To find our other newsletter, follow this link.
- Rough Guide to Preparing Software for Exascale - Introduction This Rough Guide is a Crib Sheet for improving the efficiency of parallel software for future supercomputers, and was last updated 17th February, 2023 by g.pringle@epcc.ed.ac.uk. This guide first appeared as an appendix of a CompBioMed deliverable [1]. This guide was then updated and appeared as an appendix for the CoE EXCELERAT deliverable [2].…
- CompBioMed e-Seminar #29 - CompBioMed’s 29th e-seminar took place at on 18 January 2023, titled “OpenMP in the Exascale Era”. Modern supercomputer nodes now contain more than 100 cores, and often several GPUs. This means that the “MPI only” approach to parallel programming is coming under increasing strain and can no longer deliver the maximum performance from the hardware.…
