Software Project
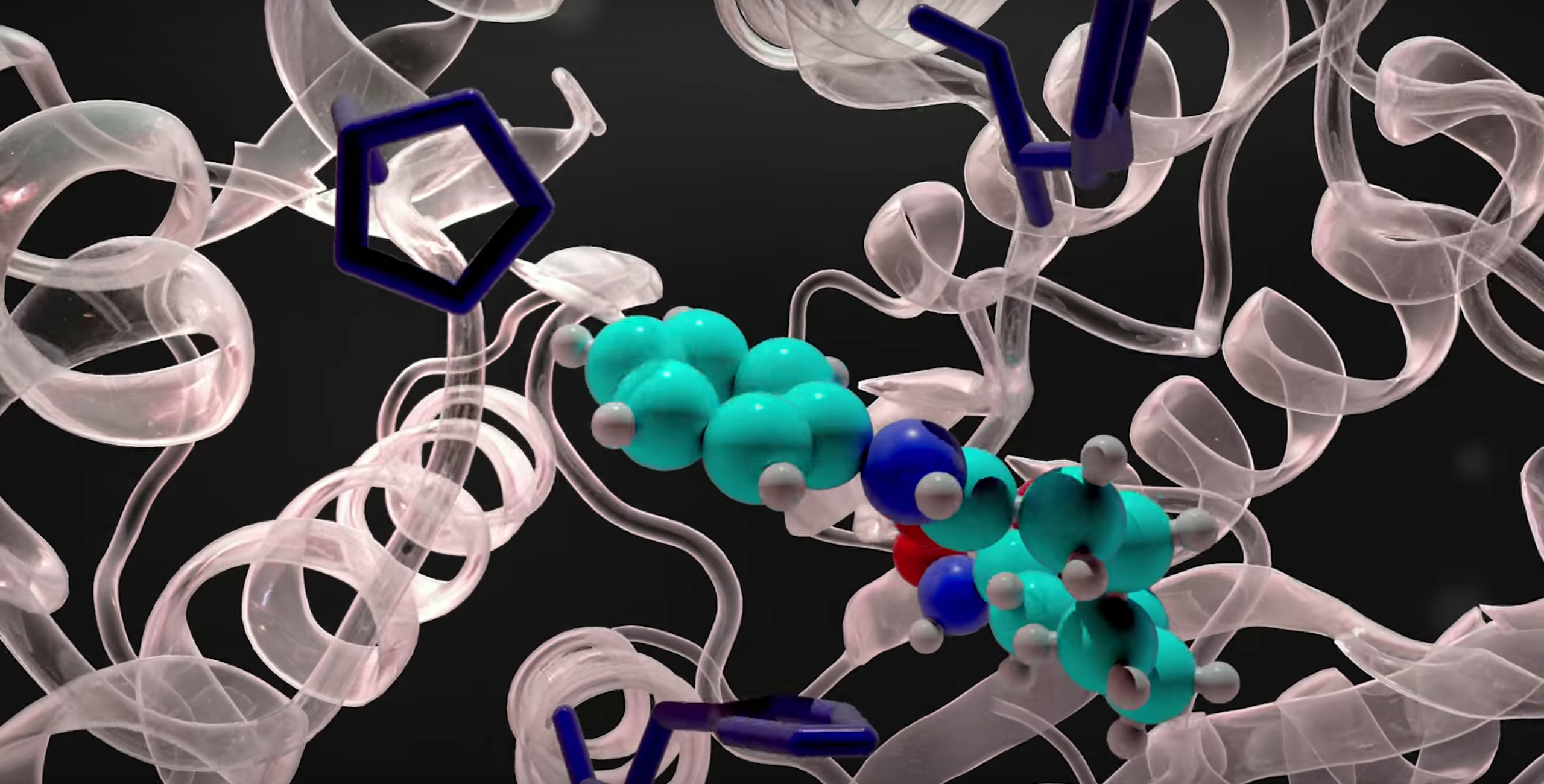
The Binding Affinity Calculator (BAC), developed by the team of Prof Peter Coveney at University College London (UK), is a workflow tool that runs and analyses simulations designed to assess how well drugs bind to their target proteins and the impact of changes to those proteins. It is a collection of scripts which wrap around common molecular dynamics codes to facilitate free energy calculations. Use of ensemble simulations to robust, accurate and precise free energy computations from both alchemical and end-point analysis methodologies. BAC is a fairly complex tool to use, so at the moment the development team at UCL have made it available as part of consulting services or research collaborations. However, EnsembleMD provides user-friendly interfaces to related binding affinity calculation services, which will be made available as an App in the on-line store of associate partner DNAnexus; a beta version is being used by pharma.
Offered By
Use scenario
Non-clinical research; Drug discovery; Design & optimization.
HPC motivation
Solve unreducible model; performs uncertainty quantification.
Relevant links
Related Articles
- S. K. Sadiq, D. Wright, S. J. Watson, S. J. Zasada, I. Stoica, Ileana, and P. V. Coveney, Automated Molecular Simulation-Based Binding Affinity Calculator for Ligand-Bound HIV-1 Proteases, Journal of Chemical Information and Modeling, 48, (9), 1909-1919, (2008), DOI: 10.1021/ci8000937.
- A. Bhati, S. Wan, D. Wright, P. V. Coveney, Rapid, accurate, precise and reliable relative free energy prediction using ensemble based thermodynamic integration, Journal of Chemical Theory and Computation, 13 (1), 210–222 (2017), DOI: 10.1021/acs.jctc.6b00979
- D. Wright, B. Hall, O. Kenway, S. Jha, P. V. Coveney, Computing Clinically Relevant Binding Free Energies of HIV-1 Protease Inhibitors, Journal of Chemical Theory and Computation, (2014), 10 (3), 1228–1241 DOI: 10.1021/ct4007037
 |
 |

