The use of existing pharmaceutical to treat COVID-19 has several potential benefits over the development of new drugs from the group up, not least that much shorter clinical trials would be needed and production on industrial scales has already been realised in many cases. Andrew Potterton, a PhD student at UCL and Evotec, described to us some of the work his research group, along with members of the Centre for Computational Science at UCL are carrying out.
The group are running binding free energy calculations for a wide range of known compounds, targeting the SARS-CoV-2 protein, with the aim of aiding drug-discovery efforts. The small-molecule/peptide pharmaceuticals they are investigating will be needed to treat the disease in the short-term until a vaccine is developed, which is expected to be approximately one year away. There are two strands to the research they are carrying out. The first strand consists of calculations to prediction the binding affinities of the approved small-molecule pharmaceuticals which could be quickly repurposed to treat COVID-19. The second strand is to use research findings to provide binding free energies of fragments as training data for a machine learning docking protocol developed by Argonne National laboratory (outlined in our previous blog post and video in our webinar series).
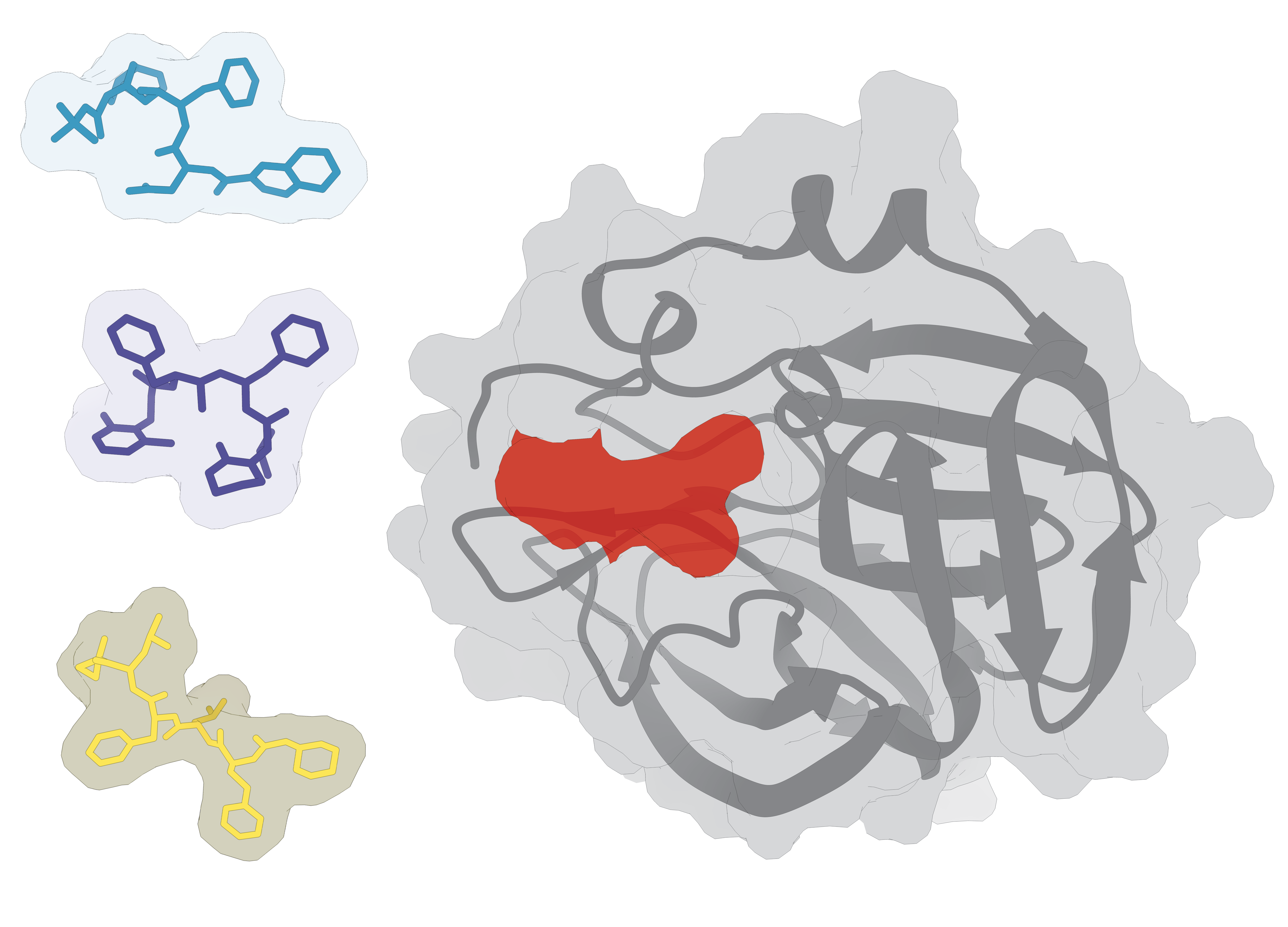
Both strands are targeting four of the viral proteins: the main protease (3CL-protease), the papain-like protease (PL-protease), the ADP-ribose-1-phosphate (NSP-3), and the endonuclease (NSP-15). In all cases, they are targeting the active site or the substrate binding site. Calculations are already running using ESMACS (Enhanced Sampling of Molecular dynamics with Approximation of Continuum Solvent) simulations on SuperMUC-NG, one of the top-10 most powerful supercomputers in the world, to screen out poor binders. Future calculations will involve running more rigorous alchemical TIES (Thermodynamic Integration with Enhanced Sample) simulations.
The main-protease of SARS-CoV-2 with 3 potential small-molecules that are approved for other targets.